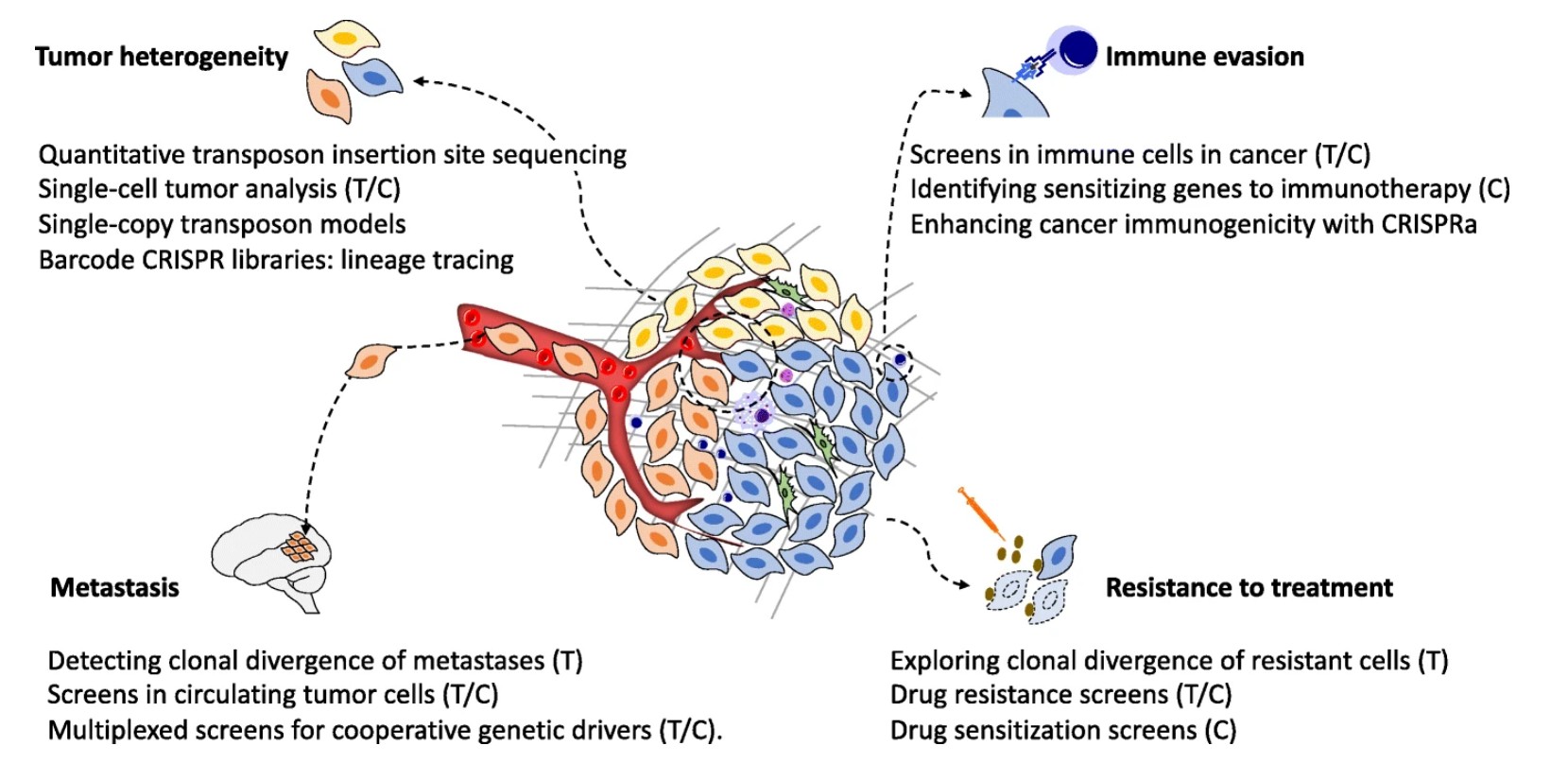
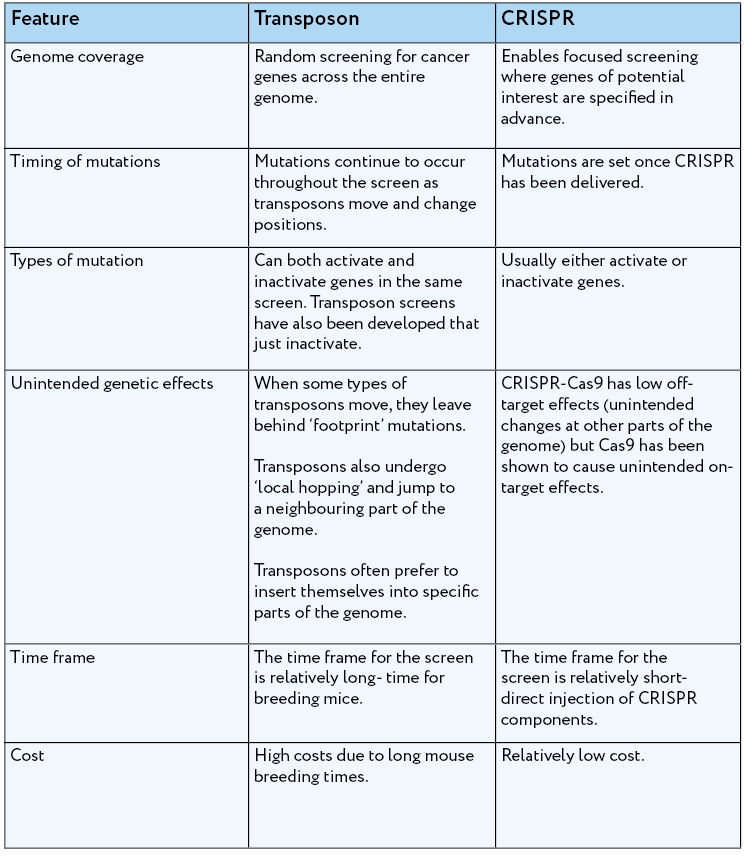
What can we use cancer screens for?
Cancers can be very different, even within the same tumour, making it difficult to identify the genes that are driving the cancer and to develop effective targeted treatments. As a result, researchers try to identify and target ‘truncal drivers’, the ancestral mutations that occur early on in a cancer’s family tree and so are likely to be present in many cancer cell types in the tumour. Transposon and CRISPR cancer screens have been developed and used to successfully identify truncal drivers of cancer. In particular, transposon screens have identified new tumour suppressor genes that cooperate with the gene Pten and act as drivers of prostate cancer.
Metastasis, when cancer cells break away from the original site and settle in other parts of the body, is one of the leading causes of cancer-related deaths. Metastatic or advanced prostate cancer is responsible for almost all deaths from prostate cancer. The spread of cancer is dependent on a complex network of genes which are not fully understood. Transposon and CRISPR cancer screens have been used to explore the genetic interactions that lead to the metastasis of multiple cancer types. Dr Jorge de la Rosa’s PCRC project will investigate the growth and spread of prostate cancer using CRISPR-Cas9.
Transposon and CRISPR screens have also been used to explore the genetics behind therapy resistance. Unfortunately, many cancers can develop resistance to treatment caused by new mutations in the cancer cells which enable them to survive in the presence of that treatment. One well-known example is the development of resistance to hormone therapy in prostate cancer cells. The genome of advanced cancers is usually very complex as they have acquired lots of genetic changes during their lifetime. This can make it difficult to identify the genes that drive resistance to treatment.
Most potentially cancerous cells are recognised and destroyed by the immune system at early stages, preventing them from developing into cancer. However, some cancers are able to avoid detection or shut down immune system components this is known as immune escape. Genetic cancer screens can be used to identify the genes that are important for the immune response which could then be used as new drug targets for immunotherapy.